summary & projects
approach
Our work integrates the fields of genomics, neurobiology, and stem cells, defining the disrupted molecular mechanisms that contribute to brain disease, in order to better facilitate the clinical translation of genetic findings. Our focus lies in resolving the convergence of, and complex interplay between, the many risk variants linked to brain disease.
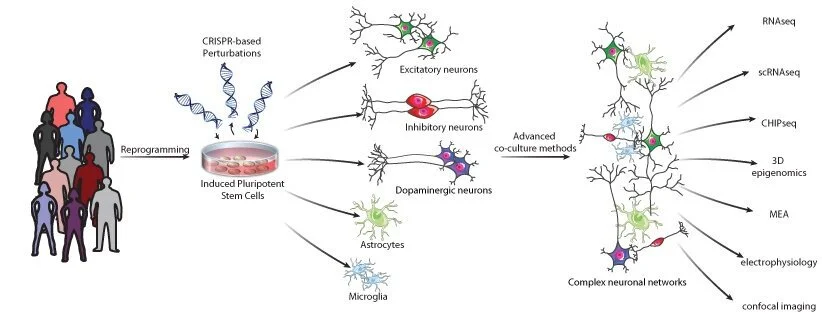
There is an urgent need for functional validation of risk variants in a cell-type-specific and context-dependent manner, in order to facilitate the clinical translation of genetic findings. We apply a functional genomics-driven approach to explore the impact of risk variants associated with brain disease, in order to improve diagnostics, predict clinical trajectories, and identify pathways that could serve as pre-symptomatic points of therapeutic intervention. The goal of our research is to advance disease modelling by integrating human induced pluripotent stem cell (hiPSC)-based approaches with multiplexed CRISPR-mediated (epi)genomic engineering, resolving the combinatorial impact of patient-specific variants across donors, cell types, and environmental contexts. This will allow us to uncover disease-associated interactions between the cell types of the brain, querying the non-cell autonomous impact(s) of complex genetic risk within increasingly sophisticated neuronal circuits. Our highly collaborative strategy integrates genetic and genomic approaches with functional analyses, towards understanding the convergence and synergy of disease risk variants within and between the cell types of the brain.
Figure 1. The Brennand laboratory integrates stem cell approaches and CRISPR-based genomic and epigenomic editing to functionally resolve how genetic variants, as well as the interactions between variants and cell types, impact cellular function and underly risk to neuropsychiatric disease.
projects
1. Establishing human induced pluripotent stem cells of neuropsychiatric disorders. We pioneered a novel approach to study complex genetic disorders such as schizophrenia (Nature, 2011; Molecular Psychiatry, 2015), bipolar disorder (Nature, 2015), and autism spectrum disorder (Molecular Psychiatry, 2016), through the reprogramming of skin samples from patients into human induced pluripotent stem cells (hiPSCs), and their subsequent differentiation into disease-relevant cell types.
2. Functional validation of the impact of common variants associated with risk for psychiatric disease. We observed remarkable concordance of genomic signals between genetic, post-mortem, and hiPSC-based studies of psychiatric disease (Nature Communications, 2017; Nature Communications, 2018; Science, 2018). We highlighted the cell-type-specific effects of common variants and demonstrated an unexpected synergistic effect between schizophrenia risk genes that converged on synaptic function (Nature Genetics, 2019a; Nature Protocols, 2021).
3. Evaluation of the mechanisms through which rare variants impact neuronal function. Highly penetrant, disease-associated rare variants are frequently pleiotropic and associated with heterogeneous clinical diagnosis, severity, and prognosis. We reported that in a genotype-dependent manner, the phenotypic impact of patient-specific NRXN1+/- mutations could occur through a reduction in wild-type NRXN1α isoform levels as well as, unexpectedly, the presence of mutant NRXN1α isoforms (Nature Genetics, 2019b).
recent research papers
1. Wang M†, Li A†, Sekiya M†, Beckmann ND†, Quan X†, Schrode N, Fernando MB, Yu A, Zhu L, Cao J, Lyu L, Yu A, Horgusluoglu E, Wang Q, Guo L, Wang Y-s, Neff R, Song W-m, Wang E, Shen Q, Zhou X, Ming C, Ho S-M, Vatansever S, Kaniskan HU, Jin J, Zhou M-M, Ando K, Ho L, Slesinger PA, Yue Z, Zhu J, Katsel P, Gandy S, Ehrlich ME, Fossati V, Noggle S, Cai D, Haroutunian V, Iijima KM#, Schadt ES#, Brennand KJ#, Zhang B#. Molecular Networks and Key Regulators of the Dysregulated Neuronal System in Alzheimer's Disease. Neuron S0896-6273(20)30861-8. doi: 10.1016/j.neuron.2020.11.002. Online ahead of print. #Co-corresponding authors.
2. Flaherty E*, Zhu S*, Barretto N, Cheng E, Deans PJM, Fernando MB, Schrode N, Francoeur N, Antoine A, Alganem K, Halpern M, Deikus G, Shah H, Fitzgerald M, Ladran I, Gochman P, Rapoport J, Tsankova NM, McCullumsmith R, Hoffman GE, Sebra R, Fang G#, Brennand KJ.# 2019. Neuronal impact of patient-specific aberrant NRXN1α splicing. Nature Genetics. 51(12):1679-1690. PMID: 31784728 . #Co-corresponding authors.
3. Schrode N*, Ho SM*, Yamamuro K, Dobbyn A, Huckins L, Matos MR, Cheng E, Deans PJM, Flaherty E, Barretto N, Topol A, Alganem K, Abadali S, Gregory J, Hoelzli E, Phatnani H, Singh V, Girish D, Aronow B, Mccullumsmith R, Hoffman GE, Stahl EA, Morishita H, Sklar P, Brennand KJ. 2019. Functional evaluation of common variant genes associated with schizophrenia. Nature Genetics. 51(10):1475-1485. doi: 10.1038/s41588-019-0497-5. PMID: 31548722. PMCID: PMC6778520.
4. Rajarajan P*, Borrman T*, Liao W*, Schrode N, Casiño C, Ho S-M, Flaherty E, Espeso-Gil S, Hoffman G.E., Hahn C-G, Weng Z#, Brennand KJ#, Akbarian S#. 2018. Neuron-specific signatures in the chromosomal connectome are associated with schizophrenia risk. Science. 362(6420). pii: eaat4311. doi: 10.1126/science.aat4311. PMID: 30545851. PMCID: PMC6408958. #Co-corresponding authors.
5. Readhead B, Hartley BJ, Eastwood BJ, Collier DA, Evans D, Farias R, He C, Hoffman G, Sklar P, Dudley JT, Schadt EE*, Savic R*, Brennand KJ*. 2018. Expression-based drug screening of neural progenitor cells from individuals with schizophrenia. Nature Communications, 9:4412. PMID: 30356048. PMCID: PMC6200740. *Co-corresponding authors.
6. Hoffman GE*, Hartley BJ, Flaherty E, Ladran I, Gochman P, CommonMind Consortium, Ruderfer D, Stahl EA, Rapoport J, Sklar P, Brennand KJ*. 2017. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. BioRxiv and Nature Communications. 8(1):2225. doi: 10.1038/s41467-017-02330-5. PMID: 29263384. PMCID: PMC5738408. *Co-corresponding authors.
recent reviews
1. Townsley, KG, Brennand KJ#, Huckins LM#. 2020. Massively parallel techniques for cataloguing the regulome of the human brain. Nature Neuroscience. 23(12):1509-1521. doi: 10.1038/s41593-020-00740-1. Epub 2020 Nov 16. #Co-corresponding authors.
2. Fernando MB, Ahfeldt T, Brennand KJ. 2020. Using stem cells to model complex genetic architectures of neuropsychiatric disease. Nature Genetics. 52(4):363-369. doi: 10.1038/s41588-020-0596-3. PMID: 32203467
3. Rehbach K, Fernando M, Brennand KJ. 2020. Potentials and challenges of human disease modeling in defined hiPSC derived 2D systems. Journal of Neuroscience. 40(6):1176-1185. doi: 10.1523/JNEUROSCI.0518-19.2019. PMID: 32024766. PMCID: PMC7002154.
4. Hoffman GE, Schrode N, Flaherty E and Brennand KJ. 2019. New considerations for hiPSC-based models of neuropsychiatric disorders. Molecular Psychiatry. 24(1):49-66. doi: 10.1038/s41380-018-0029-1. PMID: 29483625. PMCID: PMC6109625.